Aminoacyl-tRNA synthetases (aaRSs)
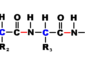
aminoacyl-tRNA synthetases (aaRSs) When ribosomes pair a tRNA (transfer ribonucleic acid) with a codon (three bases in a DNA). An amino acid is expected to be carried by the tRNA. Since each tRNA is matched with its amino acid before it meets the ribosome. The ribosome has no way of knowing if the match was made. The match is made by a family of enzymes called aminoacyl-tRNA synthetases. These enzymes charge each tRNA with the proper amino acid via a covalent ester bond. While allowing each tRNA to make the proper translation from the genetic code of DNA into the amino acid code of proteins. Cells make at least 20 different aminoacyl-tRNS synthetases, one for each of the amino acids.
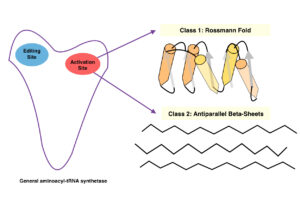
Aminoacyl-tRNA synthetases classes
Aminoacyl-tRNA synthetases belong to two classes, depending on which amino acid they specify. Class I enzymes usually are monomeric and attach to the carobxyl of their specific amino acid to the 2’ OH of adenosine 76 in the tRNA molecule. Class II enzymes are either dimeric or tetrameric and attach to their amino acids at the 3’ OH. These enzymes catalyze first by activating the amino acid by forming an aminoacyl-adenylate. Here the carboxyl of the amino acid is linked to the alpha-phosphate of ATP, displacing pyrophosphate. After the corrected tRNA is bound, the aminoacyl group of the aminoacyl- adenylate is transferred to the 2’ or 3’ terminal OH of the tRNA.
Recent studies have shown that aminoacyl-tRNA synthetases can tell the difference between the right and the wrong tRNA before they ever start catalysis. And if the enzyme binds aminoacyl-adenylate first, it is even more specific during tRNA binding.
Previous studies have also proved that aminoacyl-tRNA synthetases reject wrong tRNAs during catalysis. Other research has shown that specific aaRSs play roles in cellular fidelity, tRNA processing, RNA splicing, RNA trafficking, apoptosis, and transcriptional and translational regulation. These new revelations may present new evolutionary models for the development of cells and perhaps opportunities for pharmaceutical advancements.